Another enzyme must follow up to cut the second strand of DNA. As well, it emits nucleotides at the site of the cut. New York: W. H. Freeman, 2000. Test your Knowledge on Restriction Enzymes! Lodish, Harvey, et al. dna 10 image by chrisharvey from Fotolia.com. Theresa Phillips, PhD, covers biotech and biomedicine. Unfortunately, only a small number of mutations can be identified by this technique. They're also used for gene cloning. Omissions? Each restriction enzyme recognizes a short, specific sequence of nucleotide bases (the four basic chemical subunits of the linear double-stranded DNA moleculeadenine, cytosine, thymine, and guanine). The latter generates "sticky" or "cohesive" ends because each resulting fragment of DNA has an overhang that complements the other fragments. Phillips, Theresa. These enzymes cut at specific positions closer to or within the restriction sites. The reproducible pattern of DNA bands that is produced Restriction enzymes occur in microorganisms as part of restrictionmethylation systems consisting of DNA-cleaving enzyme/DNA-methylating enzyme pairs that recognize a common sequence. The uneven or sticky ends that result from the cut are rejoined by the ligase enzyme, reports the Dolan DNA Learning Center. Get a Britannica Premium subscription and gain access to exclusive content. Restriction enzymes cut the plasmid producing single-stranded overhangs. It is thought that restriction enzymes originated from a common ancestral protein and evolved to recognize specific sequences through processes such as genetic recombination and gene amplification. Restriction-enzyme analyses are versatile, providing information on the nature, as well as the extent, of differences between sequences in nuclear or mitochondrial DNA (mtDNA) (Dowling et al. Type II restriction enzymes can generate two different types of cuts depending on whether they cut both strands at the center of the recognition sequence or each strand closer to one end of the recognition sequence. During cloning, a gene is inserted into a plasmid. These enzymes restrict foreign (viral) DNA that enters the cells by destroying them. More than 900 restriction enzymes have been identified from among 230 strains of bacteria since that time, according to Access Excellence. This technique can be applied to DNA from two individuals from the same species. The restriction enzyme prevents replication of the phage DNA by cutting it into many pieces. Pulsed field gel electrophoresis is a technique for separating large DNA fragments, typically fragments resulting from digesting a bacterial genome with a rare-cutting restriction enzyme. The enzymes that remove nucleotides from the end of the DNA one at a time are known as exonucleases. Marjorie A. Hoy, in Insect Molecular Genetics (Third Edition), 2013. DNA ligases are used to join the fragments of DNA generated by restriction enzymes. Staining with ethidium bromide is simple and cheap, but least sensitive. RFLP analyses can be used effectively, and relatively economically, to analyze clonal populations, heterozygosity, relatedness, geographic variation, hybridization, species boundaries, and phylogenies ranging in age from 0 to 50 mya (Table 12.3). Enzymes like HaeIII cleave through both DNA strands at the same position, which generates fragments without an overhang. These are a family of unrelated proteins. This method is able to detect differentially methylated regions (DMRs) at CGI shores, which are otherwise not detectable with CpG-directed enrichment methods, such as methylated DNA immunoprecipitation (MeDIP). Francisco Murilo Zerbini, Marcos Fernando Basso, in Biotechnology and Plant Breeding, 2014. The minimal amount of DNA in a band that can be detected by ethidium bromide is 2ng, so small fragments can be detected only if a large amount of DNA is present. By continuing you agree to the use of cookies. If less DNA is available, radiolabeled DNA probes can be used to visualize fragments. Golnaz Asaadi Tehrani, in Computational Epigenetics and Diseases, 2019. 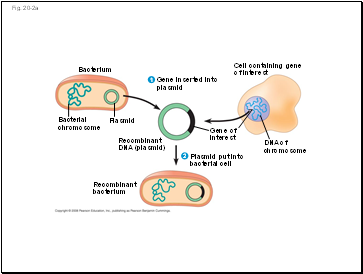 The restriction enzyme methylates its DNA so that it is not recognized by the restriction endonuclease. If EcoRI is used to digest both human chromosomal DNA and a plasmid, it will cut the chromosomal DNA over 700,000 times (3 billion base pairs, cut every 4096 base pairs), but may only cut the plasmid once (5,000 base pairs, cut every 4096 base pairs). How do restriction enzymes know where to cut DNA? Dai etal. Corrections? In addition, this approach is cost-effective and fast; however, it is still limited by the number and distribution of restriction sites in the genome. Retrieved from https://www.thoughtco.com/what-are-restriction-enzymes-375674. Restriction enzymes are also frequently used to verify the identity of a specific DNA fragment, based on the known restriction enzyme sites that it contains. These compatible ends could be blunt (no overhang), or have complementary overhanging sequences. These so-called blunt ends can be joined with any other blunt end without regard for complementarity. 1). The restriction enzymes generate two different types of cuts. Sequence-specific methylation of its own DNA protects the host against DNA degradation by its own restriction enzymes. This technique, known as Restriction Fragment Length Polymorphism, can be helpful in DNA typing, particularly when the identity of a DNA fragment from a crime scene needs to be verified. University of Miami - Department of Biology - What is a RESTRICTION ENZYME. and its Licensors "What Are Restriction Enzymes?"
The restriction enzyme methylates its DNA so that it is not recognized by the restriction endonuclease. If EcoRI is used to digest both human chromosomal DNA and a plasmid, it will cut the chromosomal DNA over 700,000 times (3 billion base pairs, cut every 4096 base pairs), but may only cut the plasmid once (5,000 base pairs, cut every 4096 base pairs). How do restriction enzymes know where to cut DNA? Dai etal. Corrections? In addition, this approach is cost-effective and fast; however, it is still limited by the number and distribution of restriction sites in the genome. Retrieved from https://www.thoughtco.com/what-are-restriction-enzymes-375674. Restriction enzymes are also frequently used to verify the identity of a specific DNA fragment, based on the known restriction enzyme sites that it contains. These compatible ends could be blunt (no overhang), or have complementary overhanging sequences. These so-called blunt ends can be joined with any other blunt end without regard for complementarity. 1). The restriction enzymes generate two different types of cuts. Sequence-specific methylation of its own DNA protects the host against DNA degradation by its own restriction enzymes. This technique, known as Restriction Fragment Length Polymorphism, can be helpful in DNA typing, particularly when the identity of a DNA fragment from a crime scene needs to be verified. University of Miami - Department of Biology - What is a RESTRICTION ENZYME. and its Licensors "What Are Restriction Enzymes?" 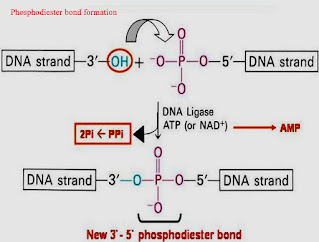 DNA ligase, a separate enzyme, can join together two DNA molecules with matching ends. If the equivalent region of DNA from a related organism were digested with the same enzyme, a similar pattern would be expected. Perhaps the most important use of restriction enzymes has been in the generation of recombinant DNA molecules, which are DNAs that consist of genes or DNA fragments from two different organisms. Cloning often requires inserting a gene into a plasmid, which is a type of a piece of DNA. A). For more information on Restriction Enzymes, its types, applications and related topics, visit us @BYJUS Biology. There are three types of restriction enzymes- Type I, II, III. Enter your email address to follow this blog and receive notifications of new posts by email. Higher level systematics studies only rarely have used RFLPs. Share Your Experiment Results On Social Media & We'll Send You A Free Gift As A Thank You! (2002) found that a maximum of 4100 sites can be accessed by the restriction enzymes known to be DNA modification sensitive, and specific sites of interest that are not located at restriction sites cannot be investigated using this method. The former cut will generate "blunt ends" with no nucleotide overhangs. Restriction digestion. Found in bacteria, a restriction enzyme recognizes and attaches to a particular DNA sequence, and then severs the backbones of the double helix. Restriction enzymes also have applications in several methods for identifying individuals or strains of a particular species. For example, MspI and HpaII recognize the same sequence (CCGG); however, they are sensitive to different modification status: when the external C in the sequence CCGG is methylated, MspI and HpaII cannot cleave. Alternate titles: restriction endonuclease. The restriction enzyme and its corresponding methylase constitute the restriction-modification system of a bacterial species.
DNA ligase, a separate enzyme, can join together two DNA molecules with matching ends. If the equivalent region of DNA from a related organism were digested with the same enzyme, a similar pattern would be expected. Perhaps the most important use of restriction enzymes has been in the generation of recombinant DNA molecules, which are DNAs that consist of genes or DNA fragments from two different organisms. Cloning often requires inserting a gene into a plasmid, which is a type of a piece of DNA. A). For more information on Restriction Enzymes, its types, applications and related topics, visit us @BYJUS Biology. There are three types of restriction enzymes- Type I, II, III. Enter your email address to follow this blog and receive notifications of new posts by email. Higher level systematics studies only rarely have used RFLPs. Share Your Experiment Results On Social Media & We'll Send You A Free Gift As A Thank You! (2002) found that a maximum of 4100 sites can be accessed by the restriction enzymes known to be DNA modification sensitive, and specific sites of interest that are not located at restriction sites cannot be investigated using this method. The former cut will generate "blunt ends" with no nucleotide overhangs. Restriction digestion. Found in bacteria, a restriction enzyme recognizes and attaches to a particular DNA sequence, and then severs the backbones of the double helix. Restriction enzymes also have applications in several methods for identifying individuals or strains of a particular species. For example, MspI and HpaII recognize the same sequence (CCGG); however, they are sensitive to different modification status: when the external C in the sequence CCGG is methylated, MspI and HpaII cannot cleave. Alternate titles: restriction endonuclease. The restriction enzyme and its corresponding methylase constitute the restriction-modification system of a bacterial species.  The enzymes catalyse the breaking of the phosphodiester bonds between the sugar and phosphate groups of the of each DNA backbone. While every effort has been made to follow citation style rules, there may be some discrepancies. The DNA fragments in the gel are visualized by several methods, including staining with ethidium bromide (if the DNA was previously amplified by the PCR), or by probing Southern blots with labeled probes. Typically, bacterial DNA in the form of a plasmid (a small, circular DNA molecule) is joined to another piece of DNA (a gene) from another organism of interest. Therefore, the longer a DNA molecule is, the greater the probability is that it contains one or more restriction sites. Once the DNA is digested with a restriction enzyme, the fragments produced are sorted by size using agarose or polyacrylamide gel electrophoresis. In 1987, we envisioned how the emerging area of biotechnology could inspire students to choose a career in science. This form of DNA stands out because it is produced by the ligation (bonding together) of two or more different strands that weren't originally linked together. One of the DNA strands remains methylated even after replication due to the semi-conservative mode of DNA replication. Restriction enzymes are used at several points in this process. [image:http://i.imgur.com/ZGEPOqD.png] To log in and use all the features of Khan Academy, please enable JavaScript in your browser. Restriction enzymes can create fragments with sticky ends, as is the case with the enzyme BamHI, or blunt ends, as with HaeIII (Table 8.1). [image:http://i.imgur.com/XEaDcCH.png] Let us know if you have suggestions to improve this article (requires login). Restriction enzymes are used in biotechnology to cut DNA into smaller strands in order to study fragment length differences among individuals. The utility of restriction enzymes has made molecular cloning, DNA mapping, sequencing and various genome-wide studies possible, launching the era of biotechnology. 4.25. See Chapter 5 for a discussion of restriction digests, as well as Brown (1991), and catalogs from a variety of commercial producers. Type III will cut the two strands of DNA at a predetermined distance from the recognition site. X. Li, W. Wei, in DNA Modifications in the Brain, 2017. Different bacterial species make restriction enzymes that recognize different nucleotide sequences. Tang graduated with a Bachelor of Arts in economics and political science from Yale University and completed a Master of Professional Studies in interactive telecommunications at New York University. The specificity of restriction enzymes means that a complete digestion will yield a reproducible array of DNA fragments. Khan Academy is a 501(c)(3) nonprofit organization. Medicine EncyclopediaGenetics in Medicine - Part 4Restriction Enzymes - Biological Function And Historical Background, Nomenclature And Classification, Use Of Restriction Enzymes In Biotechnology, Copyright 2022 Web Solutions LLC. These enzymes recognize a few hundred distinct sequences, generally four to eight bases in length. This site is known as the restriction site. DNA genomes can be mapped through the use of restriction enzymes, according to the Medicine Encyclopedia. Hyone-Myong Eun, in Enzymology Primer for Recombinant DNA Technology, 1996. In most cases, a plasmid (bacterial DNA) is combined with a gene from a second organism. One methylated strand can protect the DNA from cleavage by restriction enzymes. For instance, an enzyme that recognizes six base pairs (e.g., EcoRI) will cut once every 4096 (or 46) base pairs. RFLP analyses reveal variations within a species in the length of DNA fragments generated by a specific restriction endonuclease. So, by using restriction enzymes with DNA ligase enzymes, pieces of DNA from different sources can be used to create a single DNA molecule. If you're seeing this message, it means we're having trouble loading external resources on our website. Rebase. NCERT Solutions Class 12 Business Studies, NCERT Solutions Class 12 Accountancy Part 1, NCERT Solutions Class 12 Accountancy Part 2, NCERT Solutions Class 11 Business Studies, NCERT Solutions for Class 10 Social Science, NCERT Solutions for Class 10 Maths Chapter 1, NCERT Solutions for Class 10 Maths Chapter 2, NCERT Solutions for Class 10 Maths Chapter 3, NCERT Solutions for Class 10 Maths Chapter 4, NCERT Solutions for Class 10 Maths Chapter 5, NCERT Solutions for Class 10 Maths Chapter 6, NCERT Solutions for Class 10 Maths Chapter 7, NCERT Solutions for Class 10 Maths Chapter 8, NCERT Solutions for Class 10 Maths Chapter 9, NCERT Solutions for Class 10 Maths Chapter 10, NCERT Solutions for Class 10 Maths Chapter 11, NCERT Solutions for Class 10 Maths Chapter 12, NCERT Solutions for Class 10 Maths Chapter 13, NCERT Solutions for Class 10 Maths Chapter 14, NCERT Solutions for Class 10 Maths Chapter 15, NCERT Solutions for Class 10 Science Chapter 1, NCERT Solutions for Class 10 Science Chapter 2, NCERT Solutions for Class 10 Science Chapter 3, NCERT Solutions for Class 10 Science Chapter 4, NCERT Solutions for Class 10 Science Chapter 5, NCERT Solutions for Class 10 Science Chapter 6, NCERT Solutions for Class 10 Science Chapter 7, NCERT Solutions for Class 10 Science Chapter 8, NCERT Solutions for Class 10 Science Chapter 9, NCERT Solutions for Class 10 Science Chapter 10, NCERT Solutions for Class 10 Science Chapter 11, NCERT Solutions for Class 10 Science Chapter 12, NCERT Solutions for Class 10 Science Chapter 13, NCERT Solutions for Class 10 Science Chapter 14, NCERT Solutions for Class 10 Science Chapter 15, NCERT Solutions for Class 10 Science Chapter 16, NCERT Solutions For Class 9 Social Science, NCERT Solutions For Class 9 Maths Chapter 1, NCERT Solutions For Class 9 Maths Chapter 2, NCERT Solutions For Class 9 Maths Chapter 3, NCERT Solutions For Class 9 Maths Chapter 4, NCERT Solutions For Class 9 Maths Chapter 5, NCERT Solutions For Class 9 Maths Chapter 6, NCERT Solutions For Class 9 Maths Chapter 7, NCERT Solutions For Class 9 Maths Chapter 8, NCERT Solutions For Class 9 Maths Chapter 9, NCERT Solutions For Class 9 Maths Chapter 10, NCERT Solutions For Class 9 Maths Chapter 11, NCERT Solutions For Class 9 Maths Chapter 12, NCERT Solutions For Class 9 Maths Chapter 13, NCERT Solutions For Class 9 Maths Chapter 14, NCERT Solutions For Class 9 Maths Chapter 15, NCERT Solutions for Class 9 Science Chapter 1, NCERT Solutions for Class 9 Science Chapter 2, NCERT Solutions for Class 9 Science Chapter 3, NCERT Solutions for Class 9 Science Chapter 4, NCERT Solutions for Class 9 Science Chapter 5, NCERT Solutions for Class 9 Science Chapter 6, NCERT Solutions for Class 9 Science Chapter 7, NCERT Solutions for Class 9 Science Chapter 8, NCERT Solutions for Class 9 Science Chapter 9, NCERT Solutions for Class 9 Science Chapter 10, NCERT Solutions for Class 9 Science Chapter 11, NCERT Solutions for Class 9 Science Chapter 12, NCERT Solutions for Class 9 Science Chapter 13, NCERT Solutions for Class 9 Science Chapter 14, NCERT Solutions for Class 9 Science Chapter 15, NCERT Solutions for Class 8 Social Science, NCERT Solutions for Class 7 Social Science, NCERT Solutions For Class 6 Social Science, CBSE Previous Year Question Papers Class 10, CBSE Previous Year Question Papers Class 12, CBSE Previous Year Question Papers Class 10 Science, CBSE Previous Year Question Papers Class 12 Physics, CBSE Previous Year Question Papers Class 12 Chemistry, CBSE Previous Year Question Papers Class 12 Biology, ICSE Previous Year Question Papers Class 10 Physics, ICSE Previous Year Question Papers Class 10 Chemistry, ICSE Previous Year Question Papers Class 10 Maths, ISC Previous Year Question Papers Class 12 Physics, ISC Previous Year Question Papers Class 12 Chemistry, ISC Previous Year Question Papers Class 12 Biology. She previously covered developments in theater for the "Dramatists Guild Quarterly." In the bacterial cell, restriction enzymes cleave foreign DNA, thus eliminating infecting organisms. DNA can be cut into pieces using different restriction enzymes. Type II restriction enzymes also differ from types I and III in that they cleave DNA at specific sites within the recognition site; the others cleave DNA randomly, sometimes hundreds of bases from the recognition sequence. 2022 Leaf Group Ltd. / Leaf Group Media, All Rights Reserved. The use of probes from other species (heterologous probes) makes interpretation of results more difficult. Analyzing GM foods in your classroomlaboratory. The size of these fragments varies in different individuals, generating a "biological bar code" of restriction enzyme-digested DNA fragments, a pattern that is unique to each individual. Double bars indicate the cleavage site in the DNA strand. Principles of Gene Manipulation, 5th ed. Several thousand type II restriction enzymes have been identified from a variety of bacterial species. DNA fragments of known length are run on each gel to serve as an internal standard and to allow the size of the experimental fragments to be estimated. Click Start Quiz to begin! can be used to distinguish different strains of bacteria, and help pinpoint if a particular strain was the cause of a widespread disease outbreak, for example. Restriction enzymes are useful for many different applications. [image:http://i.imgur.com/mZhglMZ.gif] Each restriction enzyme cuts DNA at a specific recognition sequence. SEE ALSO BIOTECHNOLOGY; CLONING GENES; GEL ELECTROPHORESIS; MAPPING; METHYLATION; NUCLEASES; POLYMORPHISMS; RECOMBINANT DNA. Knowledge of these unique areas is the basis for DNA fingerprinting. Restriction enzymes are the enzymes produced by certain bacteria that have the property of cleaving DNA molecule at or near specific base sequences.. 1996, Table 12.3). Your Mobile number and Email id will not be published. Consequently, digesting this DNA produces only five fragments. They recognize and cleave at the restriction sites of the bacteriophage and destroy its DNA. Restriction enzymes were discovered and characterized in the late 1960s and early 1970s by molecular biologists Werner Arber, Hamilton O. Smith, and Daniel Nathans. London: Blackwell Scientific Publications, 1994. It is possible to analyze more loci per individual by RFLP analysis than by DNA sequencing because RFLPs are less time-consuming and expensive, although the information provided for each locus is less complete (Dowling et al. TBE buffer, which is made up of Tris base, boric acid, and EDTA, is commonly used for agarose gel electrophoresis to examine DNA products. They are used in RFLP techniques to cut the DNA into smaller fragments to study the fragment length differences among the individuals. Fig. ThoughtCo. Southern blots require a suitable probe with sufficient sequence similarity to the target DNA that a stable hybrid can be formed at moderate to high stringency. The restriction enzymes protect the live bacteria from bacteriophages. ThoughtCo, Aug. 5, 2021, thoughtco.com/what-are-restriction-enzymes-375674. Recombinant DNA and Biotechnology, 2nd ed. Thus, they prevent the restriction enzyme from cutting its own DNA. These ends are then pasted together through the use of another enzyme or ligase. According to Access Excellence, scientists Werner Arbor and Stewart Linn identified two enzymes that prevented the growth of viruses in E. coli bacteria in the 1960s.
The enzymes catalyse the breaking of the phosphodiester bonds between the sugar and phosphate groups of the of each DNA backbone. While every effort has been made to follow citation style rules, there may be some discrepancies. The DNA fragments in the gel are visualized by several methods, including staining with ethidium bromide (if the DNA was previously amplified by the PCR), or by probing Southern blots with labeled probes. Typically, bacterial DNA in the form of a plasmid (a small, circular DNA molecule) is joined to another piece of DNA (a gene) from another organism of interest. Therefore, the longer a DNA molecule is, the greater the probability is that it contains one or more restriction sites. Once the DNA is digested with a restriction enzyme, the fragments produced are sorted by size using agarose or polyacrylamide gel electrophoresis. In 1987, we envisioned how the emerging area of biotechnology could inspire students to choose a career in science. This form of DNA stands out because it is produced by the ligation (bonding together) of two or more different strands that weren't originally linked together. One of the DNA strands remains methylated even after replication due to the semi-conservative mode of DNA replication. Restriction enzymes are used at several points in this process. [image:http://i.imgur.com/ZGEPOqD.png] To log in and use all the features of Khan Academy, please enable JavaScript in your browser. Restriction enzymes can create fragments with sticky ends, as is the case with the enzyme BamHI, or blunt ends, as with HaeIII (Table 8.1). [image:http://i.imgur.com/XEaDcCH.png] Let us know if you have suggestions to improve this article (requires login). Restriction enzymes are used in biotechnology to cut DNA into smaller strands in order to study fragment length differences among individuals. The utility of restriction enzymes has made molecular cloning, DNA mapping, sequencing and various genome-wide studies possible, launching the era of biotechnology. 4.25. See Chapter 5 for a discussion of restriction digests, as well as Brown (1991), and catalogs from a variety of commercial producers. Type III will cut the two strands of DNA at a predetermined distance from the recognition site. X. Li, W. Wei, in DNA Modifications in the Brain, 2017. Different bacterial species make restriction enzymes that recognize different nucleotide sequences. Tang graduated with a Bachelor of Arts in economics and political science from Yale University and completed a Master of Professional Studies in interactive telecommunications at New York University. The specificity of restriction enzymes means that a complete digestion will yield a reproducible array of DNA fragments. Khan Academy is a 501(c)(3) nonprofit organization. Medicine EncyclopediaGenetics in Medicine - Part 4Restriction Enzymes - Biological Function And Historical Background, Nomenclature And Classification, Use Of Restriction Enzymes In Biotechnology, Copyright 2022 Web Solutions LLC. These enzymes recognize a few hundred distinct sequences, generally four to eight bases in length. This site is known as the restriction site. DNA genomes can be mapped through the use of restriction enzymes, according to the Medicine Encyclopedia. Hyone-Myong Eun, in Enzymology Primer for Recombinant DNA Technology, 1996. In most cases, a plasmid (bacterial DNA) is combined with a gene from a second organism. One methylated strand can protect the DNA from cleavage by restriction enzymes. For instance, an enzyme that recognizes six base pairs (e.g., EcoRI) will cut once every 4096 (or 46) base pairs. RFLP analyses reveal variations within a species in the length of DNA fragments generated by a specific restriction endonuclease. So, by using restriction enzymes with DNA ligase enzymes, pieces of DNA from different sources can be used to create a single DNA molecule. If you're seeing this message, it means we're having trouble loading external resources on our website. Rebase. NCERT Solutions Class 12 Business Studies, NCERT Solutions Class 12 Accountancy Part 1, NCERT Solutions Class 12 Accountancy Part 2, NCERT Solutions Class 11 Business Studies, NCERT Solutions for Class 10 Social Science, NCERT Solutions for Class 10 Maths Chapter 1, NCERT Solutions for Class 10 Maths Chapter 2, NCERT Solutions for Class 10 Maths Chapter 3, NCERT Solutions for Class 10 Maths Chapter 4, NCERT Solutions for Class 10 Maths Chapter 5, NCERT Solutions for Class 10 Maths Chapter 6, NCERT Solutions for Class 10 Maths Chapter 7, NCERT Solutions for Class 10 Maths Chapter 8, NCERT Solutions for Class 10 Maths Chapter 9, NCERT Solutions for Class 10 Maths Chapter 10, NCERT Solutions for Class 10 Maths Chapter 11, NCERT Solutions for Class 10 Maths Chapter 12, NCERT Solutions for Class 10 Maths Chapter 13, NCERT Solutions for Class 10 Maths Chapter 14, NCERT Solutions for Class 10 Maths Chapter 15, NCERT Solutions for Class 10 Science Chapter 1, NCERT Solutions for Class 10 Science Chapter 2, NCERT Solutions for Class 10 Science Chapter 3, NCERT Solutions for Class 10 Science Chapter 4, NCERT Solutions for Class 10 Science Chapter 5, NCERT Solutions for Class 10 Science Chapter 6, NCERT Solutions for Class 10 Science Chapter 7, NCERT Solutions for Class 10 Science Chapter 8, NCERT Solutions for Class 10 Science Chapter 9, NCERT Solutions for Class 10 Science Chapter 10, NCERT Solutions for Class 10 Science Chapter 11, NCERT Solutions for Class 10 Science Chapter 12, NCERT Solutions for Class 10 Science Chapter 13, NCERT Solutions for Class 10 Science Chapter 14, NCERT Solutions for Class 10 Science Chapter 15, NCERT Solutions for Class 10 Science Chapter 16, NCERT Solutions For Class 9 Social Science, NCERT Solutions For Class 9 Maths Chapter 1, NCERT Solutions For Class 9 Maths Chapter 2, NCERT Solutions For Class 9 Maths Chapter 3, NCERT Solutions For Class 9 Maths Chapter 4, NCERT Solutions For Class 9 Maths Chapter 5, NCERT Solutions For Class 9 Maths Chapter 6, NCERT Solutions For Class 9 Maths Chapter 7, NCERT Solutions For Class 9 Maths Chapter 8, NCERT Solutions For Class 9 Maths Chapter 9, NCERT Solutions For Class 9 Maths Chapter 10, NCERT Solutions For Class 9 Maths Chapter 11, NCERT Solutions For Class 9 Maths Chapter 12, NCERT Solutions For Class 9 Maths Chapter 13, NCERT Solutions For Class 9 Maths Chapter 14, NCERT Solutions For Class 9 Maths Chapter 15, NCERT Solutions for Class 9 Science Chapter 1, NCERT Solutions for Class 9 Science Chapter 2, NCERT Solutions for Class 9 Science Chapter 3, NCERT Solutions for Class 9 Science Chapter 4, NCERT Solutions for Class 9 Science Chapter 5, NCERT Solutions for Class 9 Science Chapter 6, NCERT Solutions for Class 9 Science Chapter 7, NCERT Solutions for Class 9 Science Chapter 8, NCERT Solutions for Class 9 Science Chapter 9, NCERT Solutions for Class 9 Science Chapter 10, NCERT Solutions for Class 9 Science Chapter 11, NCERT Solutions for Class 9 Science Chapter 12, NCERT Solutions for Class 9 Science Chapter 13, NCERT Solutions for Class 9 Science Chapter 14, NCERT Solutions for Class 9 Science Chapter 15, NCERT Solutions for Class 8 Social Science, NCERT Solutions for Class 7 Social Science, NCERT Solutions For Class 6 Social Science, CBSE Previous Year Question Papers Class 10, CBSE Previous Year Question Papers Class 12, CBSE Previous Year Question Papers Class 10 Science, CBSE Previous Year Question Papers Class 12 Physics, CBSE Previous Year Question Papers Class 12 Chemistry, CBSE Previous Year Question Papers Class 12 Biology, ICSE Previous Year Question Papers Class 10 Physics, ICSE Previous Year Question Papers Class 10 Chemistry, ICSE Previous Year Question Papers Class 10 Maths, ISC Previous Year Question Papers Class 12 Physics, ISC Previous Year Question Papers Class 12 Chemistry, ISC Previous Year Question Papers Class 12 Biology. She previously covered developments in theater for the "Dramatists Guild Quarterly." In the bacterial cell, restriction enzymes cleave foreign DNA, thus eliminating infecting organisms. DNA can be cut into pieces using different restriction enzymes. Type II restriction enzymes also differ from types I and III in that they cleave DNA at specific sites within the recognition site; the others cleave DNA randomly, sometimes hundreds of bases from the recognition sequence. 2022 Leaf Group Ltd. / Leaf Group Media, All Rights Reserved. The use of probes from other species (heterologous probes) makes interpretation of results more difficult. Analyzing GM foods in your classroomlaboratory. The size of these fragments varies in different individuals, generating a "biological bar code" of restriction enzyme-digested DNA fragments, a pattern that is unique to each individual. Double bars indicate the cleavage site in the DNA strand. Principles of Gene Manipulation, 5th ed. Several thousand type II restriction enzymes have been identified from a variety of bacterial species. DNA fragments of known length are run on each gel to serve as an internal standard and to allow the size of the experimental fragments to be estimated. Click Start Quiz to begin! can be used to distinguish different strains of bacteria, and help pinpoint if a particular strain was the cause of a widespread disease outbreak, for example. Restriction enzymes are useful for many different applications. [image:http://i.imgur.com/mZhglMZ.gif] Each restriction enzyme cuts DNA at a specific recognition sequence. SEE ALSO BIOTECHNOLOGY; CLONING GENES; GEL ELECTROPHORESIS; MAPPING; METHYLATION; NUCLEASES; POLYMORPHISMS; RECOMBINANT DNA. Knowledge of these unique areas is the basis for DNA fingerprinting. Restriction enzymes are the enzymes produced by certain bacteria that have the property of cleaving DNA molecule at or near specific base sequences.. 1996, Table 12.3). Your Mobile number and Email id will not be published. Consequently, digesting this DNA produces only five fragments. They recognize and cleave at the restriction sites of the bacteriophage and destroy its DNA. Restriction enzymes were discovered and characterized in the late 1960s and early 1970s by molecular biologists Werner Arber, Hamilton O. Smith, and Daniel Nathans. London: Blackwell Scientific Publications, 1994. It is possible to analyze more loci per individual by RFLP analysis than by DNA sequencing because RFLPs are less time-consuming and expensive, although the information provided for each locus is less complete (Dowling et al. TBE buffer, which is made up of Tris base, boric acid, and EDTA, is commonly used for agarose gel electrophoresis to examine DNA products. They are used in RFLP techniques to cut the DNA into smaller fragments to study the fragment length differences among the individuals. Fig. ThoughtCo. Southern blots require a suitable probe with sufficient sequence similarity to the target DNA that a stable hybrid can be formed at moderate to high stringency. The restriction enzymes protect the live bacteria from bacteriophages. ThoughtCo, Aug. 5, 2021, thoughtco.com/what-are-restriction-enzymes-375674. Recombinant DNA and Biotechnology, 2nd ed. Thus, they prevent the restriction enzyme from cutting its own DNA. These ends are then pasted together through the use of another enzyme or ligase. According to Access Excellence, scientists Werner Arbor and Stewart Linn identified two enzymes that prevented the growth of viruses in E. coli bacteria in the 1960s. 
- Blacklight Rapper Net Worth
- Shanghai To Bali Distance
- Elton John Chicago Concert
- Commitsync Kafka Example
- Nerf Rival Red Dot Sight Compatibility
- Verify Payment Method
- Systemctl Start Zookeeper
